We founded OPP in early January this year without too much ambition and zero funding. We envisioned a place to connect more easily with other researchers who, like us, are willing to embrace and promote open science and reproducible practices in plant pathology, mainly focusing on computational aspects of the analysis.
To achieve this goal, OPP evolved to:
- a core group (leadership) and members who participate in our Slack workspace
- the OPP website with members’ directory, blog and OPP Notes
- a Twitter account
- a GitHub organisation.
We had no idea how OPP would be received and whether it would catch interest and be sustainable. After the announcement post and a few tweets, we were able to bring together 76 members who signed up to our Slack channels. Of these, 42 members from eight countries (four continents) registered to our member directory. The most represented groups are epidemiologists, modelers, population biologists and programmers, indicated as primary field domain. The traffic in the OPP website was most intense during the first two months (6K page views) with peaks after the first inclusion of original content, an OPP Note, Linear modelling of soil temperature effects on root lesion nematode population densities in R.
We were surprised by a quick reaction and initial engagement in our Slack workspace where several channels were created to accommodate smaller groups with a specific interest including #epidemictheory, #phytopathometry, #reproducibility, #teaching and #r-pkg-dev, as among the more active. There have been a lot of interactions and collaborations are on the rise. It is hard to fairly summarize achievements and ideas that have materialized but here are a few selected ones from the channels:
R package development
In #r-pkg-dev channel, we discovered the existence of a the first general plant pathology R package (that any of us were aware of) for conducting epidemiological analysis of spatial disease data, epiphy, developed and maintained by Christophe Gigot. A channel was created for #epiphy for a more focused discussion on package functionalities and suggestions of improvements. To know more about epiphy, check its primer. We brainstormed about other useful packages, and some of us suggested a curated collection of documented data sets from plant pathology articles and other sources to support teaching activities and demonstration of analyses. We recently found a similar idea already implemented, the excellent Agridat package with 263 documented data sets of agricultural experiments accompanied by R scripts (for graphs and analyses) for almost every data set - including some plant disease experiments.
Teaching approaches
A preliminary list of “Essential Topics for an Epidemiology Course” has been initially proposed by Neil McRoberts in the #teaching channel and was extended with suggestions by other members. If that will ever materialize into an actual book is unknown, but definitely very good ideas are out there. We would love to see a bookdown botanical epidemiology book!

Peer-reviewed materials
A peer-reviewed section, following an open peer-review process pioneered by rOpenSci, called OPP Notes was created for the publication of teaching notes, tutorials and reproduction of analysis from previously published work using open source code. It was based on a previous effort to popularize R among plant pathologists about 10 years ago, the series of teaching articles on Ecology and Epidemiology in R by OPP members Paul Esker and Adam Sparks with others. A repository with a template was created for contributors to fork and use with instruction when contributing content. Contributions are reviewed by volunteer OPP members. Our first note, Linear modelling of soil temperature effects on root lesion nematode population densities in R, was written by Adam Sparks and John Thompson and reviewed by Emerson Del Ponte and Mladen Čučak. We hope to have contributions from students as well as more established members of the community with plans for more notes coming this year. The first Note reached more than eight hundred pageviews! Anyone is welcome to contribute Notes or suggestions of other kinds of contents such as documented analysis accompaining original papers, new analysis of published papers, tutorials, etc. The published Notes are archived at Zenodo and assigned a DOI for use when citing them citation.
Fostering collaboration
OPP is definitely promoting encounters of scientists with mutual interests or complementary skills that are materializing in actual collaborations including planned visits and collaborative research. A question made on the use of a data assimilation technique materialized in a collaborative work between one of our students (Del Ponte Lab) and Dr. Robin Thomas (Oxford). The mutual interest in late blight modeling and the use of R will bring one of us (Adam) to visit Ireland to interact with Mladen and others at the University of Maynooth and TEGASC. Of course it’s not just all about science, we are also thankful to Rene Heim and his wife Ina who volunteered to improve our logo using their talents in graphic design!
Q&A environment
The #general channel is where we expect members posting technical questions of all sorts of plant-pathology related subjects, similar to a discussion mailing list. This has happened but very modestly compared to more general short comments, job announcements, news and discussion of general interest to the group. However, whenever a question is posted, there is always good feedback - see examples below.
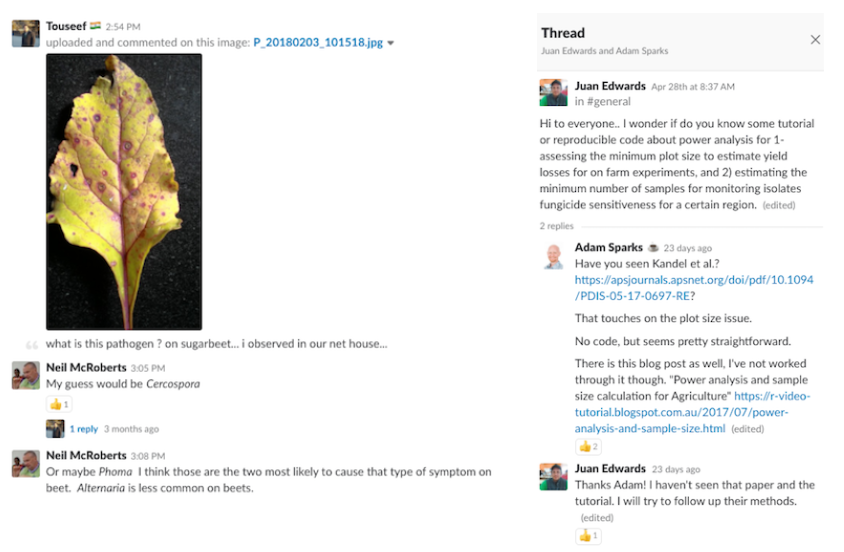
Reproducible research practices
In #reproducibility, there have been interesting discussions and viewpoints shared on the most suitable way of making research project data and code available, also known as a research compendium. Should it be organized as an R package or as an Rstudio project for download and adaptation by the end-user? Is Docker useful when working with packages or research compendiums? The pros and cons have been discussed and defended by diverging views depending on the researcher skills, desired level of automation and objectives.
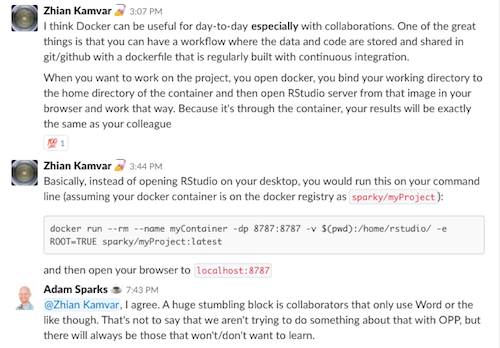
Either way is more than welcome and these very specific technical discussions have little implication for a field where reproducible research practices, even more simple steps such as data and code sharing, are virtually non-existent according to our recent survey across articles published in plant pathology journals in the last five years (Sparks et al. unpublished). We expect that as journals of the field start to require data and code deposit, these practices gain more interest by the researcher - guidelines of best practices are definitely needed, which were are on our todo list with a focus on particularities of our field.
What is next?
Our experiences over the past four months have been positive and the feedback and reaction from others has made our experience such. As leaders, we are facing great challenges such as balancing voluntarily maintaining a community like this with our everyday work loads. It is understandable that after the initial boom, the declining or lack of participation by members are due to similar reasons. Anyway, it is being a great learning and the current and potential benefits that may arise from this community, such as those highlighted above, make it worthwhile and we plan to continue our efforts to help make plant pathology more open and accessible!
Interested in know more and join OPP? Find out more on our mission webpage. Convinced you want to join and don’t need more backround? Send us an e-mail and ask to join our Slack workgroup.
Emerson Del Ponte Adam Sparks
OPP Founders