Dr. Susann Auer, a postdoc at Technische Universität Dresden (Dresden, Germany), delivered the first OPP Virtual Seminar on the 31st of March, entitled Molecular response of Clubroot infected plants to the endophytic fungus Acremonium alternatum. I had the privilege to get to know her and her work a little better. Dr. Auer is a passionate researcher, who shows care towards her students and is thoughtful in considering her fellow researchers.
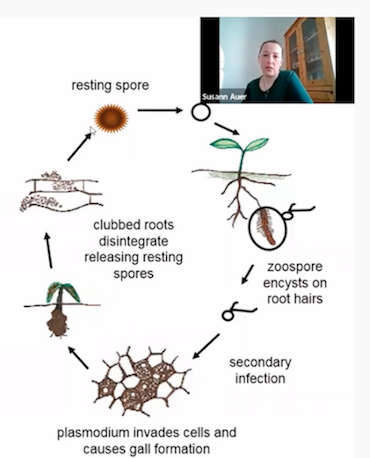
The first discovery or description of Clubroot disease was in the 19th century. Why do you think it is so poorly understood to date? I can see, it is not a lack of trying because the group you belong alone has been doing this research for at least 10 years.
Yes, Clubroot or club root, depending on your training, was discovered in the 19th century. You can read more about it on our groups webpage. I think one of the most important things to know is that the causal organism of Clubroot disease, Plasmodiophora brassicae is neither a fungus nor an oomycete, it is a protist, with a biphasic life cycle. We know much more about Clubroot now than we did 15 years or so ago. I think it is still poorly understood because of a few reasons (know more).
- Funding is limited - reviewers of grants (and papers) often don’t grasp the importance and economic damage of that disease in Brassica crops.
- It is a tricky pathogen to work with - the experiments take a long time, and you need a good understanding of the basic biology of the disease before you can design proper experiments.
- There is a low number of research groups working with Clubroot currently compared to other diseases.
- The first genome draft of one pathotype (of many) was only available in 2015, from another genome draft the original data are still not available.
- Clubroot groups are often competing for first papers and do not collaborate as well as they probably could.
- Lack of trained scientists in Clubroot disease - as far as I see it many Clubroot Principle Investigators are well in their 50s or older, and there are only few early career researchers working on this disease, this poses a challenge because we need to make sure knowledge can be transferred between these two groups.
In the last years Clubroot research was facilitated mainly by Canadian researchers that were equipped with public funds for clubroot research since rapeseed production is very important for their economy. Proper funding of well-thought out research accelerates knowledge and gives researchers the freedom and time they need to study specific aspects of the disease.
Our group leader has studied the disease since her PhD and publishes a lot on Clubroot. She has collaborated with many groups in the past and continues to do this which definitely helped a lot as well - but our funds are limited. We are a small group, with only her and me (and the occasional student) working on Clubroot in the recent years. We now have a new postdoc, who started doing some work on Clubroot as well. As we know, when starting with a new lab group and a new disease (especially this challenging pathogen) you need to gain experience. However, we have industry projects that tackle small things - and here we often cannot publish our results, due to the nature of industry. Overall, companies see that Clubroot is a rising problem and start investing resources by funding small projects because they are desperate now. Many countries often still ignore this problem, in the EU there is no concerted effort yet to tackle this disease. I fear we will only receive good funding when it’s pretty much too late and the majority of Brassica fields are already infested.
I think it is important to share with students that laboratory work is not always what you expect. What extraction protocol do you use for the RNA from roots? and how do you find its efficiency?
I used various methods, reagents and kits, depending on the host plant. For Arabidopsis, I mostly use RNAzol (a liquid, not a kit), it usually gives good results. I recently started using Nucleozol which seems to work just as fine. Often, I harvest my plants, homogenize the tissue immediately and freeze it in RNAzol until I have the complete experimental set harvested. Only then do I start with the RNA extraction. This gives the best results in terms of reproducibility and RNA quality. For Brassica, which has more specialized compounds that negatively affect RNA extraction, I used the innuprep RNA kit from Jenaanalytik and my all-time favourite Qiagen RNA plant mini kit. The latter is pretty expensive though so I mostly stick with RNAzol.
Regarding efficiency it is hard to say, no matter what set of samples we analyse the variation is always considerable even if the same amount of biomass is used. From my experience, I think this interference can come from many sources. One of the first causes is the extraction efficiency of the researcher with the use of liquid nitrogen or beads, the variation usually decreases when the researcher gains experience. Some of the major sources are due to the complexity of this pathogen and its interaction with the host.
Due to the nature of this patho-system you are left with a mixed sample, the roots of the host, especially in the advanced stages of disease consist mainly of pathogenic tissue making it difficult to extract host material. Also, you never know the ratio of pathogen to plant in your tissue so a considerable amount of your extracted RNA could be pathogen RNA. So even if the quality and quantity of your whole RNA sample seems fine you could end up with a low amount of transcripts for your plant genes of interest. I think a student needs to also have an awareness of seemingly small but actually important things. The longer you store your samples in the freezer the less likely you are to have good quality RNA in the end.
In your opinion, why is it crucial to understand the mechanisms which underlie the response of the host to the pathogen at a molecular level for this specific pathosystem, and the Plasmodiophora x Acremonium interaction?
Susann: We don’t really know much about the immediate molecular response of plants to two microbes that are applied simultaneously yet. Luckily an increasing amount of papers addresses interactions with 3 or more organisms at the same time now. These interaction studies are complex and can be difficult to analyse. Especially when applying a BCA we should know which pathways are employed that might lead to the reduction of symptoms and ultimately increase yields in the end. Without this knowledge, we are poking at things in the dark, which is wasteful to resources and unwise experimental design. For new studies, we need the molecular knowledge to make educated guesses on what makes sense to try to combat a disease.
What statistical program are you using for your data analysis? If it is R, what packages do you find most useful?
Susann: For my qPCR data analyses, I use an old Excel macro - known as the REST software tool which allows for several reference genes to be included when estimating gene expression. I currently don’t use R directly for my data analysis. For visualisation, I use online tools such as BoxplotR and others to make plots. R is something I have worked with in the past, but I have had to prioritise my time and weigh up the benefits for me in the season I am in now. Luckily I have colleagues who are experienced with R and use it on a daily basis. If I have a question or problem I just ask them for help.
We are in the IYPH2020, why do you think it is important to integrate biological control agents, like Acremonium, into IPM systems?
Susann: Biocontrol agents are natural antagonists. They work because they coevolve with the pest organisms we try to combat. In this specific year of plant health, it is important to remember that we already have a lot of tools at our hands we could try out to combat Clubroot disease. In many countries people use BCAs that work well in their specific context, sometimes due to a lack of alternatives or no access to expensive chemicals. Studying the interaction of the host with a BCA equips us with the necessary knowledge needed to be able to apply more BCAs to detrimental diseases. The need for sustainable agriculture that does not harm the farmers applying pest control or the environment and diversity of species is likely the best long-term route. In the EU a lot of effective yet harmful substances are banned now so we need to investigate alternatives and we need to do it now.
In your Virtual Seminar you spoke of potential collaborations, can we have some details regarding this?
We currently collaborate with a group in Czech, that does the -omics analyses for us and with several industry partners. I would love to have a collaboration with people with experience in developing fungal species into biofertilizers or BCAs. I would also very much like to work with other fungal specialists that can help me sequence the species I investigate or provide bioinformatic knowledge.
A huge thank you to Susann for making the time to deliver her Virtual presentation, answer my questions and meet with me on Zoom. It was an honour to share your work with the OPP community. We look forward to seeing how we can grow together.
You can go and watch Susann’s Virtual Seminar on our official YouTube channel. You are also welcome to interact with us in our Slack Community on our #virtual-seminar channel. By clicking on this link you can request to Join Open Plant Pathology on Slack.