Background
Bacterial wilt pathogens in the Ralstonia solanacearum species complex (RSSC) are destructive pathogens, especially in tropical and sub-tropical climates with year-round precipitation. Most strains in this species complex are soil-borne pathogens that cause persistant infestations once introduced. Tomato, potato, tobacco, banana, ginger, and peanut are crops that are well-recognized hosts of Ralstonia. In 2005, Mark Fegan and Philippe Prior formalized a DNA-based taxonomy for the RSSC called a “phylotype-sequevar” system . Since their publication, hundreds of plant pathologists around the world have classified the population diversity of the RSSC on specific crops or in geographic regions.
What is the database for?
RSSCdb is a database of characterized strains of Ralstonia solanacearum species complex strains whose data have extracted from the peer-reviewed articles that include the geographic, host of origin, year, and phylogenetic classification (phylotype/sequevar). This strain metadata is available in the main text or supplemental material of the publication. The project was developed to provide UC Davis undergraduate researchers with a remote research experience. The data will be supplemental material for a systematic review article to be submitted for publication after data collection is finalized (Lowe-Power et al. unpublished). In the interim, the data is released on BioRxiv and GitHub after UC Davis students complete their thesis (https://www.biorxiv.org/content/10.1101/2020.07.13.189936v1).
How is it developed?
The database is organized in an Excel spreadsheet, a format that facilitates regular curation of data entry.
What’s next?
Emerson M. Del Ponte will advise a student on creating an interactive dashboard to view the data, similar to his FGSCdb for tracking species and chemotypes of the Fusarium graminearum species complex: https://fgsc.netlify.app/
We would like to extend the utility of the Ralstonia diversity project by allowing other researches to contribute raw data to populate the global map. Get in touch (tlowepower@ucdavis.edu) to receive instructions on how to submit your data.
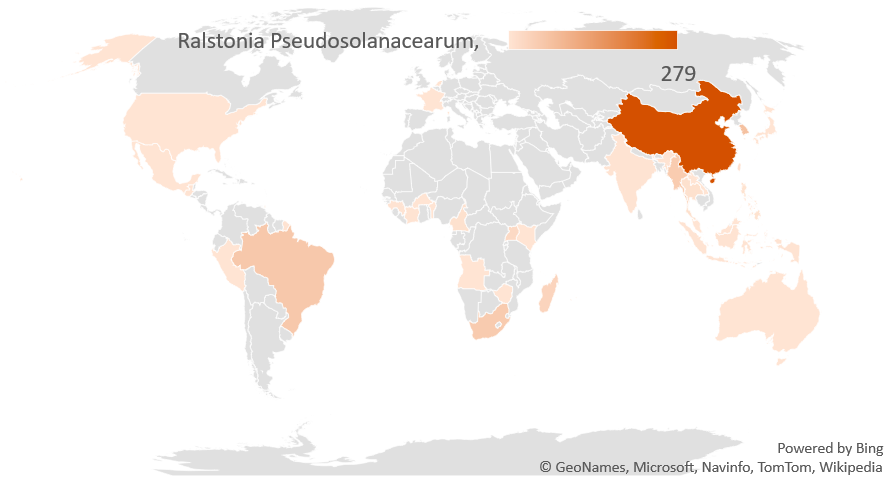
Data source: https://github.com/lowepowerlab/Ralstonia_Global_Diversity
Author and maintainer: Tiffany Lowe-Power and Emerson M. Del Ponte
Reference: Fegan, M., & Prior, P. (2005). How complex is the Ralstonia solanacearum species complex. Bacterial wilt disease and the Ralstonia solanacearum species complex, 1, 449-61.